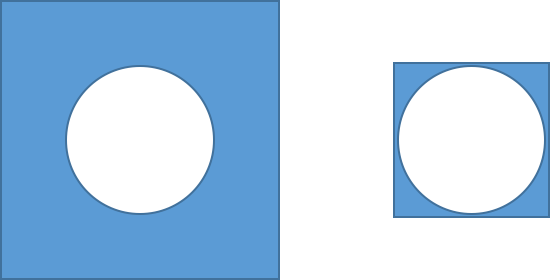
- Recall the previous circle problem. We needed a square that enclosed the circle. Which of the two situations will be more efficient?
-
Clearly the case on the right is better. We will have fewer wasted darts in the smaller blue square area on the right.
-
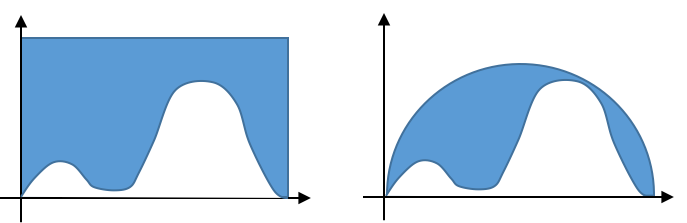
Similarly, the case on the right in the figure below will be more accurate.
-
But how do we sample uniformly under the half circle the right figure?
- For a rectangle, we just used a uniform random number generator to get random x and y points.
- There are routines for generating random points that fit some given distribution, such as normally distributed random points.
Approach
- Let the curve we want to generate points on be $g(x)$.
- Let $G(x)=\int_a^xg(x)dx,$ where $a$ is the lower bound of integration.
- We sample uniformly on $G$ and invert to find the corresponding $x=G^{-1}$.
- The resulting $x$ locations will be distributed consistent with $g(x)$.
- To get the $y$ location of a point corresponding to $x$, we sample a random number uniformly on $[0,,g(x)]$.
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
from scipy.special import erf
from scipy.optimize import fsolve
from scipy.integrate import quad
plt.rc('font', size=14)
def make_plot():
μ = 2
σ = 0.5
x = np.linspace(0,4,100)
g = 1/np.sqrt(2*np.pi*σ**2)*np.exp(-1/2/σ**2*(x-μ)**2)
G = 0.5*(1+erf(1/σ/np.sqrt(2)*(x-μ)))
fig, (ax1, ax2) = plt.subplots(1,2, figsize=(10,5))
ax1.plot(x,g)
ax1.set_xlabel('x'); ax1.set_ylabel('g(x)')
ax1.set_xlim([0,4]); ax1.set_ylim([-0.1,1])
ax2.plot(x,G)
ax2.set_xlabel('x'); ax2.set_ylabel('G(x)')
ax2.set_xlim([0,4]); ax2.set_ylim([-0.1,1])
def Ginv(xFind, Gwant):
return 0.5*(1+erf(1/σ/np.sqrt(2)*(xFind-μ))) - Gwant
for yy in np.linspace(0.01,0.99,15):
xx = fsolve(Ginv, μ, args=yy)[0]
ax2.arrow(4, yy, xx-4+0.1, 0, head_width=0.02, head_length=0.1, fc='gray', ec='gray', linestyle='dashed')
ax2.arrow(xx, yy, 0, -yy+0.05-0.1, head_width=0.05, head_length=0.05, fc='gray', ec='gray')
plt.tight_layout()
make_plot()

Example: let $g(x)$ be a normal distribution
n = 2000 # number of points
#---------- Define g(x)
μ = 2
σ = 0.5
maxG = 1.0
def g(x):
return 1/np.sqrt(2*np.pi*σ**2)*np.exp(-1/2/σ**2*(x-μ)**2)
#---------- get g(x) distributed random x points
rG = np.random.rand(n)*maxG # uniform samples on G
rx = np.empty(n) # corresponding x locations found next
def Ginv(xFind, Ggiven): # inverse function: G --> x
return 0.5*(1+erf(1/σ/np.sqrt(2)*(xFind-μ))) - Ggiven
for i, rGi in enumerate(rG):
rx[i] = fsolve(Ginv, μ, args=rGi)
#---------- get y points below g(x)
ry = np.empty(n)
for i in range(n):
ry[i] = np.random.rand()*g(rx[i])
#---------- plot results
/var/folders/_7/42qnnz6j17g26jhlhv2l5_580000gn/T/ipykernel_55618/596636519.py:19: DeprecationWarning: Conversion of an array with ndim > 0 to a scalar is deprecated, and will error in future. Ensure you extract a single element from your array before performing this operation. (Deprecated NumPy 1.25.)
rx[i] = fsolve(Ginv, μ, args=rGi)
x = np.linspace(0,2*μ,100)
fig,ax = plt.subplots(figsize=(10,5))
ax.plot(x,g(x), 'k-');
ax.plot(rx, ry, 'b.', markersize=2);
ax.plot(rx, np.random.rand(len(rx))*0.04-0.02-0.04, '.', markersize=1.0, color='red')
ax.set_xlabel('x');
ax.set_ylabel('g(x)');

Sampling nonuniformly distributed random numbers
- Most random number generators provide uniformly distributed random numbers on the domain $[0,,1]$.
- In the previous example, we saw how to sample random points that are normally distributed.
- Specifically, the selection of $x$ points will be normally distributed.
- This means that there are more $x$ points near $\mu$ where the peak is, than on the tails.
- In the above example, we also found corresponding random $y$ points.
- However, in this section, we are only interested in the nonuniformly distributed $x$ points.
- For example, plotting a histogram of the
rxpoints above gives:
plt.hist(rx, 40, rwidth=0.8);

- The previous method is difficult if the integration of $g(x)$ is complex or expensive and/or if the subsequent inversion of $G(x)$ is complex or expensive.
- Another method for sampling uniformly distributed points from a nonrectangular region is similar to the MC integration problem.
- However, we are not doing integration here, only showing another method for sampling from a nonuniform distribution.
- This method does not require integration of $g(x)$ or inversion of $G(x)$.
- We draw a bounding curve $F(x)$ that is easy to integrate and invert. This could be a rectangle.
- $F(x)$ is everywhere greater than $g(x)$.
- Then we draw an $r_x$ point from from the bounding curve, as previously described (using G(x)).
- Now we accept the sample point with probability $g(r_x)/F(r_x)$.
- This is done by sampling a random number between 0 and 1. If the random number is less than $g(r_x)/F(r_x)$, then we accept the point, otherwise we reject it.
- This approach is called the rejection method.
Example
- Sample random normally distributed variables using the rejection method.
def make_plot2():
n = 8000 # random numbers to try (not all kept)
μ = 2
σ = 0.5
def g(x):
return 1/np.sqrt(2*np.pi*σ**2)*np.exp(-1/2/σ**2*(x-μ)**2)
def F(x): # bounding function, here a simple uniform profile
return 1
#-----------------
nkeep = 0 # total number kept: incremented below
rx_all = np.empty(0) # storage for all points: expanded below
for i in range(n):
rx = np.random.rand()*2*μ
Paccept = g(rx)/F(rx)
r = np.random.rand()
if r<Paccept:
rx_all=np.append(rx_all, rx)
nkeep += 1
#-----------------
plt.hist(rx_all, 40, rwidth=0.8);
plt.xlim([0,4])
make_plot2()

Example-Poisson Process
- Poisson Processes are extremely important in probability theory.
- Used to model random processes in time and space
- Raindrop spacing, radioactive decay, eddies in turbulence, etc.
Raindrops
- Suppose it’s raining and you listen to the time spacing of droplets.
- There will be some average rate.
- But sometimes droplets will be very close together, and sometimes far apart.
Question: what is the distribution of the spacing between events; and can we model this?
- Find the PDF of intervals between events; that is $p(\Delta t_e)$.
- Assume events occur randomly in time with a mean event rate $\lambda$ (=) $s^{-1}$. That is, $\lambda$ drops per second, on average.
- Assume events are statistically independent of each other.
- The probability of getting an event in some interval $dt$ is $\lambda dt$, or $dt/T$, where $T=1/\lambda$ is the mean time between events.
- Consider the interval $(t_0,,t_0+\Delta t_e)$, with some sub-intervals:
| 1 2 3 4 |
|-----|-----|-----|-----|
| dt dt dt dt |
$\phantom{xxx}t_0\phantom{xxxxxxxxxxxxxxxxxx}t_0+\Delta t_e$
- The probability of not getting an event in sub-interval 1 is $1-\lambda dt$.
- The probability of not getting an event in sub-interval 1 and sub-interval 2 and $\ldots$ sub-interval 4 is
$$(1-\lambda dt)^4,$$
- or, for $n$ sub-intervals
$$(1-\lambda dt)^n \rightarrow (1-\lambda dt)^{\Delta t_e/dt}$$
-
Now, let $dt\rightarrow 0$.
-
$P(t_{\text{next event}}>\Delta t_e) = \lim_{dt\rightarrow 0}(1-\lambda dt)^{\Delta t_e/dt}$.
-
Now, at $dt=0$ we have $1^{\infty}$. Work on this:
-
Now, at $dt=0$ we have $0/0$, so use L’Hopital’s Rule.
$$\ln P = \lim_{dt\rightarrow 0}\frac{-\lambda \Delta t_e}{(1-\lambda dt)} = -\lambda \Delta t_e.$$
Hence, $$P(t_{\text{next event}}>\Delta t_e) = e^{-\lambda \Delta t_e}.$$ Or, $$P(t_{\text{next event}}<\Delta t_e) = 1- e^{-\lambda \Delta t_e}.$$
- This is a cumulative distribution function.
- Differentiate it to get the probability density function:
-
This is an exponential distribution.
-
So, to generate exponentially-distributed events, randomly sample $\Delta t_e$ values from this distribution.
- As above, draw uniform samples of $P(\Delta t_e)$ (the cumulative distribution function) on $[0,,1]$. Invert this function to get $\Delta t_e$.
- The inversion, is just solving the expression for $P(\Delta t_e)$ for $\Delta t_e$, where $P$ values are taken as random numbers between 0 and 1:
$$\Delta t_e = -\frac{\ln(1-P)}{\lambda}.$$
def make_plot3():
λ = 1
n = 2000
rP = np.random.rand(n)
Δt_e = -np.log(1-rP)/λ
x = np.linspace(0,8,1000)
y = λ*np.exp(-λ*x)
times = np.cumsum(Δt_e[:40])
plt.rc('font', size=14);
fig,(ax1,ax2) = plt.subplots(1,2,figsize=(10,5))
ax1.hist(Δt_e, 80, density=True, rwidth=0.8);
ax1.plot(x,y, linewidth=3);
ax1.set_xlabel(r'$\Delta t_e$');
ax1.set_ylabel(r'$p(\Delta t_e)$');
ax1.set_xlim([0,5])
ax2.plot(times, np.zeros(len(times)), '.', markersize=5)
for t in times:
ax2.axvline(x=t, ymin=0.45, ymax=0.55, linewidth=1, color='gray')
ax2.set_xlabel('time')
ax2.set_ylim([-1,1])
plt.tight_layout()
make_plot3()
