Multidimensional PDEs¶
- Parabolic PDE here: $$\frac{\partial f}{\partial t} = \alpha\frac{\partial^2 f}{\partial x^2} + \alpha\frac{\partial^2 f}{\partial y^2} + S.$$
- Create a two dimensional grid of $(i,\,j)$ points in the $x$ and $y$ directions.
- Take $S$ as constant in time.
FTCS method¶
- Explicit
- $\mathcal{O}(\Delta t),$ $\mathcal{O}(\Delta x^2),$ $\mathcal{O}(\Delta y^2).$
$$f_{i,j}^{n+1} = f_{i,j}^n + \frac{\alpha\Delta t}{\Delta x^2}(f_{i-1,j}^n-2f_{i,j}^n+f_{i+1,j}^n) + \frac{\alpha\Delta t}{\Delta y^2}(f_{i,j-1}^n-2f_{i,j}^n+f_{i,j+1}^n) + \Delta tS_{i,j}.$$
In [3]:
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import time
from IPython.display import display, clear_output
Lxy = 1.0 # domain length Lx=Ly
nTauRun = 0.2 # number of intrinsic timescale to run for
nxy = 22 # number of grid points in x and y directions
α = 1.0 # thermal diffusivity
tFac = 0.5 # timestep size factor.
tau = Lxy**2/α # diffusive timescale based on the whole domain.
tend = nTauRun*tau # run time
dxy = Lxy/(nxy-1) # grid spacing in x and y directions
dt = tFac*dxy**2*α/4 # timestep size
nt = int(np.ceil(tend/dt)) # number of timesteps
nPlt = np.ceil(1/tFac)*10 # how often to plot
f = np.zeros((nxy,nxy)) # solution array
S = np.ones((nxy,nxy)) # source term array
X,Y = np.meshgrid(np.linspace(0,Lxy,nxy), np.linspace(0,Lxy,nxy))
i = np.arange(1,nxy-1)
j = np.arange(1,nxy-1)
ix = np.ix_
for it in range(nt):
f[ix(i,j)] += α*dt/dxy**2*(f[ix(i-1,j)] - 2*f[ix(i,j)] + f[ix(i+1,j)]) + \
α*dt/dxy**2*(f[ix(i,j-1)] - 2*f[ix(i,j)] + f[ix(i,j+1)]) + \
dt*S[ix(i,j)]
if it%nPlt==0:
plt.clf()
plt.contourf(X,Y,f,levels=np.linspace(0,dt*260,21))
plt.colorbar()
plt.gca().set_aspect('equal', adjustable='box')
clear_output(wait=True)
display(plt.gcf())
time.sleep(0.1)
plt.clf()
<Figure size 640x480 with 0 Axes>
BTCS method¶
- Implicit
- $\mathcal{O}(\Delta t),$ $\mathcal{O}(\Delta x^2),$ $\mathcal{O}(\Delta y^2).$
$$f_{i,j}^{n+1} = f_{i,j}^n + \frac{\alpha\Delta t}{\Delta x^2}(f_{i-1,j}^{n+1}-2f_{i,j}^{n+1}+f_{i+1,j}^{n+1}) + \frac{\alpha\Delta t}{\Delta y^2}(f_{i,j-1}^{n+1}-2f_{i,j}^{n+1}+f_{i,j+1}^{n+1}) + \Delta tS_{i,j}.$$
- Rearrange this equation so that all $n+1$ terms are on the left-hand side (LHS) and all other terms are on the right-hand side (RHS):
- Let $$d_x = \frac{\alpha\Delta t}{\Delta x^2},$$ $$d_y = \frac{\alpha\Delta t}{\Delta y^2}.$$
- Then, $$ (d_y)f_{i,j-1}^{n+1} + (d_x)f_{i-1,j}^{n+1} + (-2d_x-2d_y-1)f_{i,j}^{n+1} + (d_x)f_{i+1,j}^{n+1} + (d_y)f_{i,j+1}^{n+1} = -f_{i,j}^n - \Delta tS_{i,j}. $$
- Or, in coefficient form: $$ (c)f_{i,j-1}^{n+1} + (u)f_{i-1,j}^{n+1} + (a)f_{i,j}^{n+1} + (u)f_{i+1,j}^{n+1} + (c)f_{i,j+1}^{n+1} = b,$$ $$c = d_y,$$ $$u = d_x,$$ $$a = -2d_x-2d_y-1,$$ $$b= -f_{i,j}^n-\Delta tS_{i,j}.$$
- Alternative: if $d_x=d_y$, divide through by by $d_x$. $$ (c)f_{i,j-1}^{n+1} + (u)f_{i-1,j}^{n+1} + (a)f_{i,j}^{n+1} + (u)f_{i+1,j}^{n+1} + (c)f_{i,j+1}^{n+1} = b,$$ $$c = 1,$$ $$u = 1,$$ $$a = -4-\frac{1}{d},$$ $$b= -\frac{f_{i,j}^n}{d}-\frac{\Delta tS_{i,j}}{d}.$$
Matrix form¶
- Our unknowns are on a two-dimensional grid.
- We need to arrange these in one-dimension for solution as a coupled linear system of equations.
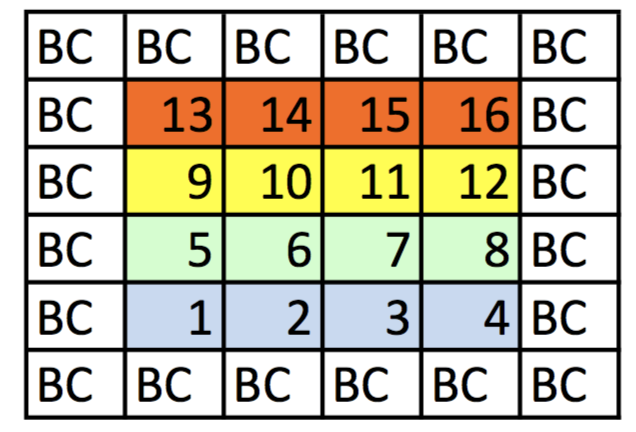
- The points in the grid are numbered sequentially.
- Three types of cells:
- Corner
- Edge
- Interior
- As for 1-D problems, unknown points (cells that are not boundary cells) will refer to neighboring cells.
- Corner and edge cells will refer to boundary cells due to the discretization stencils of the spatial derivatives in the PDE.
- These BC point will appear on the RHS of the $Ax=b$ linear system to be solved.
- Considering Dirichlet BCs.
Matrix structure:¶
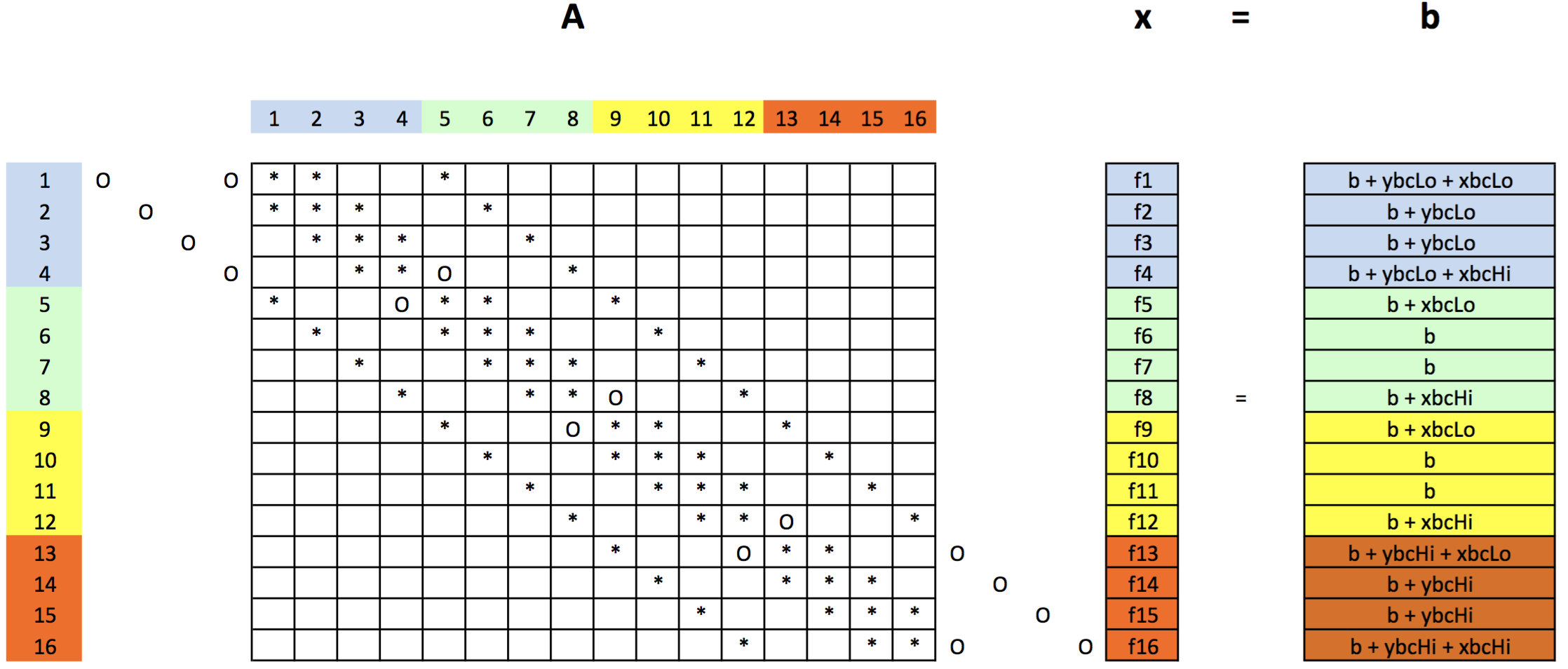
- In the matrix shown, empty cells are zero.
- Note the explicit "O" points where boundary conditons have been moved from the $A$ matrix to the RHS vector $b$.
- Note the explicit "*" points have nonzero values in the matrix.
- Consider grid point 4:
- It has neighbors: 3 and 8, so in row 4, there are *'s in columns 3, 4, and 8.
- The "O" in column 5 arises since point 5 is not a neighbor of point 4 and does not appear in point 4's discretization stencil.
- Instead, point 4's right neighbor is a boundary point, and this boundary augments the 4$^{th}$ element of the $b$ vector. Similarly for the point on the boundary below point 4.
Here is the matrix in terms of coefficients:¶
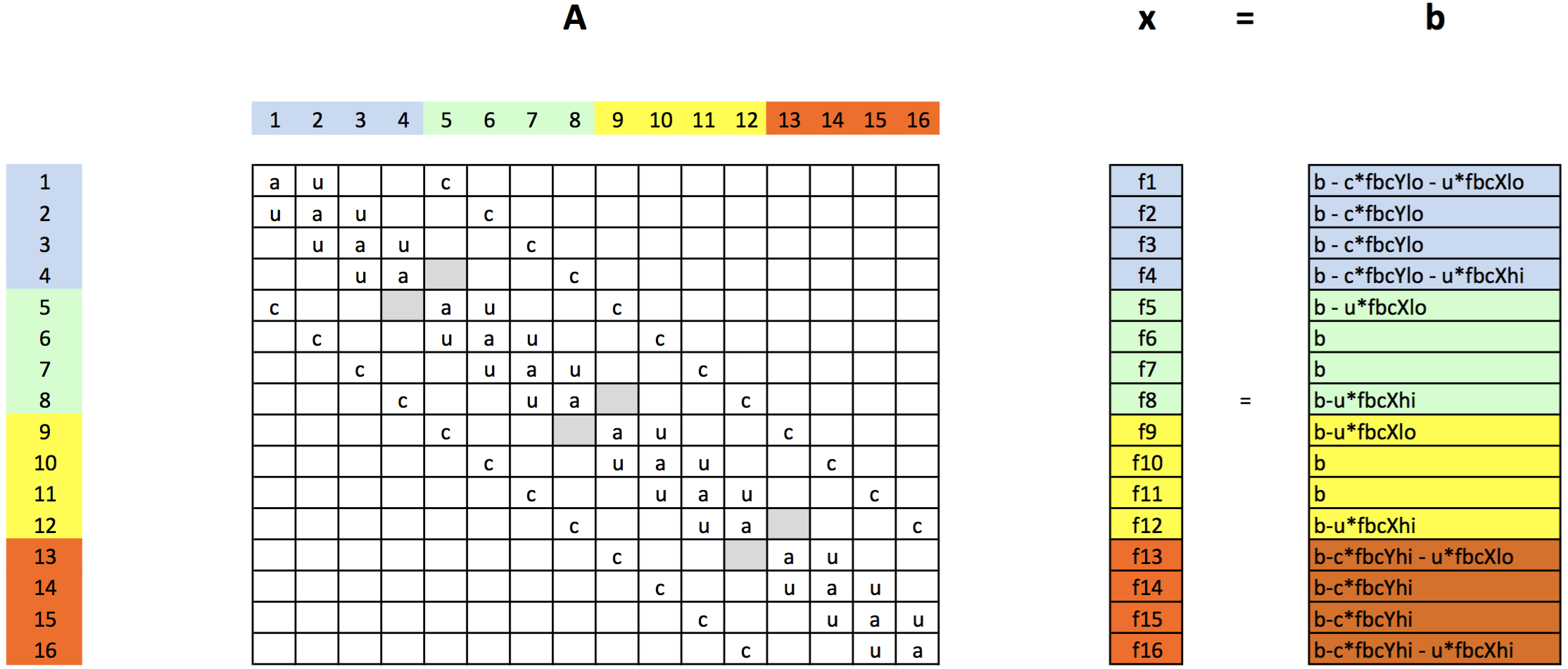
- Note, this is on a grid where the indicies of the unknowns start at 1.
- Here, point $(i,\,j)$ is unknown number $i+(j-1)\cdot n_x$, where $n_x$ is the number of points in the $x$ direction (4 in the example above).
- On a grid where the indicies of the unknowns start at 0 (as in Python), we would have point $(i,\,j)$ in the grid corresponds to point $i+j\cdot n_x$ in the list of unknowns.
In [11]:
A = np.zeros((5,5))
A[1:4,1:4] = 1
A
Out[11]:
array([[0., 0., 0., 0., 0.],
[0., 1., 1., 1., 0.],
[0., 1., 1., 1., 0.],
[0., 1., 1., 1., 0.],
[0., 0., 0., 0., 0.]])
In [12]:
A[1:4,1:4]
Out[12]:
array([[1., 1., 1.],
[1., 1., 1.],
[1., 1., 1.]])
In [14]:
i = [1,2,3]
j = [1,2,3]
A[i,j]
Out[14]:
array([1., 1., 1.])
In [7]:
i = [1,2,3]
j = [1,2,3]
A[np.ix_(i,j)]
Out[7]:
array([[1., 1., 1.],
[1., 1., 1.],
[1., 1., 1.]])
In [9]:
a = -4*np.array([1,1,1,1])
b = [1,1,1]
c = [1,1,1]
A = np.diag(a) + np.diag(b,-1) + np.diag(c,1)
In [22]:
A
Out[22]:
array([[-4, 1, 0, 0],
[ 1, -4, 1, 0],
[ 0, 1, -4, 1],
[ 0, 0, 1, -4]])
In [ ]: